

The COVID-19 outbreak simulator simulates the outbreak of COVID-19 in a population. It was first designed to simulate the outbreak of COVID-19 in small populations in enclosed environments, such as a FPSO (floating production storage and offloading vessel) but has since been expanded to simulate much larger populations with dynamic parameters.
This README file contains all essential information but you can also visit our documentation for more details. Please feel free to contact us if you would like to simulate any particular environment.
* [Background](#background) * [Basic assumptions](#basic-assumptions) * [Statistical models](#statistical-models) * [Simulation method](#simulation-method) * [Limitation of the simulator](#limitation-of-the-simulator) * [Installation and basic usage](#installation-and-basic-usage) * [Installation](#installation) * [Getting help](#getting-help) * [Command line options of the core simulator](#command-line-options-of-the-core-simulator) * [Example usages](#example-usages) * [Homogeneous and heterogeneous populations](#homogeneous-and-heterogeneous-populations) * [Change number of infectors](#change-number-of-infectors) * [Changing model parameters](#changing-model-parameters) * [Specigy group-specific parameters](#specigy-group-specific-parameters) * [Plug-in system (advanced usages)](#plug-in-system-advanced-usages) * [Specify one or more plugins from command line](#specify-one-or-more-plugins-from-command-line) * [System plugins](#system-plugins) * [Common paramters of plugins](#common-paramters-of-plugins) * [Plugin init](#plugin-init) * [Plugin dynamic-r0](#plugin-dynamic-r0) * [Plugin stat](#plugin-stat) * [Plugin sample](#plugin-sample) * [Plugin quarantine](#plugin-quarantine) * [Implementation of plugins](#implementation-of-plugins) * [Output from the simulator](#output-from-the-simulator) * [Events tracked during the simulation](#events-tracked-during-the-simulation) * [Summary report from multiple replicates](#summary-report-from-multiple-replicates) * [Data analysis tools](#data-analysis-tools) * [time_vs_size.R](#time_vs_sizer) * [merge_summary.py](#merge_summarypy) * [Acknowledgements](#acknowledgements)Background¶
Basic assumptions¶
This simulator simulates the scenario in which
- A group of individuals in a population in which everyone is susceptible. The population is by default free of virus, but seroprevalence and incidance rate could be specifed to initialize the population with virus.
- The population can be divided into multiple subgroups with different parameters.
- One or more virus carriers are introduced to the population, potentially after a fixed days of self-quarantine.
- Infectees are by default removed from from the population (separated, quarantined, hospitalized, or deceased, as long as he or she can no longer infect others) after they displayed symptoms, but options are provided to act otherwise.
- The simulation is by default stopped after the population is free of virus (all infected individuals have been removed), or everyone is infected, or after a pre-specified time (e.g. after 10 days).
- More complex simulation scenarios are provided by a plugin system where actions such as contact tracing, sampling can be simulated.
The simulator simulates the epidemic of the population with the introduction of infectors. Detailed statistics are captured from the simulations to answer questions such as:
- What is the expected day and distribution for the first person to show symptoms?
- How many people are expected to be removed once an outbreak starts?
- How effective will self-quarantine before dispatch personnels to an enclosed environment?
The simulator uses the latest knowledge about the spread of COVID-19 and is validated against public data. This project will be contantly updated with our deepening knowledge on this virus.
Statistical models¶
We developed multiple statistical models to model the incubation time, serial interval, generation time, proportion of asymptomatic transmissions, using results from multiple publications. We validated the models with empirical data to ensure they generate, for example, correct distributions of serial intervals and proporitons of asymptomatic, pre-symptomatic, and symptomatic cases.
The statistical models and related references are available at
- Statistical models of COVID-19 outbreaks (Version 1)
- Statistical models of COVID-19 outbreaks (Version 2)
The models will continuously be updated as we learn more about the virus.
Simulation method¶
This simulator simulations individuals in a population using an event-based model. Briefly speaking,
- The simulator creates an initial population with a set of initial
events(see later section for details) - The events will happen at a pre-specified time and will trigger further events. For example,
an
INFECTIONevent will cause someone to be infected with the virus. The individual will be marked asinfectedand according to individualized parameters of the infectivity of the infected individual, theINFECTIONevent may trigger a series of events such asINFECTIONto other individuals,SHOW_SYMPTIOMif he or she is symptomatic and will show symptom after an incubation period,REMOVALorQUARANTINEafter symptoms are detected. - After processing all events at a time point, the simulator will call specified plugins, which can perform a variety of operations to the population.
- The simulator processes and logs the events and stops after pre-specified exit condition.
Limitation of the simulator¶
- The simulator assumes a “mixed” population. Although some groups might be more suscepticle to the virus, people in the same group will be equally likely to be infected.
- The simulator does not simulate “contact” or “geographic locations”, so it is not yet possible to similate scenarios such as “contact centers” such as supermarkets and subways.
Installation and basic usage¶
Installation¶
This simulator is programmed using Python >= 3.6 with numpy and scipy. A conda environment is
recommended. After setting up a conda environment with Python >= 3.6 and these two packages,
please run
pip install -r requirements.txt
to install required packages, and then
pip install covid19-outbreak-simulator
to install the simulator.
Getting help¶
You can then use command
outbreak_simulator -h
to check the usage information of the basic simulator,
outbreak_simualtor --plugin -h
to get decription of common parameters for plugins, and
outbreak_simulator --plugin plugin_name -h
to get the usage information of a particular plugin.
Command line options of the core simulator¶
$ outbreak_simulator -h
usage: outbreak_simulator [-h] [--popsize POPSIZE [POPSIZE ...]]
[--susceptibility SUSCEPTIBILITY [SUSCEPTIBILITY ...]]
[--symptomatic-r0 SYMPTOMATIC_R0 [SYMPTOMATIC_R0 ...]]
[--asymptomatic-r0 ASYMPTOMATIC_R0 [ASYMPTOMATIC_R0 ...]]
[--incubation-period INCUBATION_PERIOD [INCUBATION_PERIOD ...]] [--repeats REPEATS]
[--handle-symptomatic [HANDLE_SYMPTOMATIC [HANDLE_SYMPTOMATIC ...]]]
[--pre-quarantine [PRE_QUARANTINE [PRE_QUARANTINE ...]]]
[--initial-incidence-rate [INITIAL_INCIDENCE_RATE [INITIAL_INCIDENCE_RATE ...]]]
[--initial-seroprevalence [INITIAL_SEROPREVALENCE [INITIAL_SEROPREVALENCE ...]]]
[--infectors [INFECTORS [INFECTORS ...]]] [--interval INTERVAL] [--logfile LOGFILE]
[--prop-asym-carriers [PROP_ASYM_CARRIERS [PROP_ASYM_CARRIERS ...]]]
[--stop-if [STOP_IF [STOP_IF ...]]] [--allow-lead-time] [--plugin ...] [-j JOBS]
[-s STAT_INTERVAL]
optional arguments:
-h, --help show this help message and exit
--popsize POPSIZE [POPSIZE ...]
Size of the population, including the infector that will be introduced at the beginning of
the simulation. It should be specified as a single number, or a serial of name=size values
for different groups. For example "--popsize nurse=10 patient=100". The names will be used
for setting group specific parameters. The IDs of these individuals will be nurse0, nurse1
etc.
--susceptibility SUSCEPTIBILITY [SUSCEPTIBILITY ...]
Weight of susceptibility. The default value is 1, meaning everyone is equally susceptible.
With options such as "--susceptibility nurse=1.2 patients=0.8" you can give weights to
different groups of people so that they have higher or lower probabilities to be infected.
--symptomatic-r0 SYMPTOMATIC_R0 [SYMPTOMATIC_R0 ...]
Production number of symptomatic infectors, should be specified as a single fixed number,
or a range, and/or multipliers for different groups such as A=1.2. For example "--
symptomatic-r0 1.4 2.8 nurse=1.2" means a general R0 ranging from 1.4 to 2.8, while nursed
has a range from 1.4*1.2 and 2.8*1.2.
--asymptomatic-r0 ASYMPTOMATIC_R0 [ASYMPTOMATIC_R0 ...]
Production number of asymptomatic infectors, should be specified as a single fixed number,
or a range and/or multipliers for different groups
--incubation-period INCUBATION_PERIOD [INCUBATION_PERIOD ...]
Incubation period period, should be specified as "lognormal" followed by two numbers as
mean and sigma, or "normal" followed by mean and sd, and/or multipliers for different
groups. Default to "lognormal 1.621 0.418"
--repeats REPEATS Number of replicates to simulate. An ID starting from 1 will be assinged to each replicate
and as the first columns in the log file.
--handle-symptomatic [HANDLE_SYMPTOMATIC [HANDLE_SYMPTOMATIC ...]]
How to handle individuals who show symptom, which should be "keep" (stay in population),
"remove" (remove from population), and "quarantine" (put aside until it recovers). all
options can be followed by a "proportion", and quarantine can be specified as
"quarantine_7" etc to specify duration of quarantine.
--pre-quarantine [PRE_QUARANTINE [PRE_QUARANTINE ...]]
Days of self-quarantine before introducing infector to the group. The simulation will be
aborted if the infector shows symptom before introduction. If you quarantine multiple
people or specified named groups, you will need to append the IDs to the parameter (e.g.
--pre-quarantine day nurse1 nurse2
--initial-incidence-rate [INITIAL_INCIDENCE_RATE [INITIAL_INCIDENCE_RATE ...]]
Incidence rate of the population (default to zero), which should be the probability that
any individual is currently affected with the virus (not necessarily show any symptom).
Multipliers can be specified to set incidence rate of for particular groups (e.g.
--initial-incidence-rate 0.1 docter=1.2 will set incidence rate to 0.12 for doctors).
--initial-seroprevalence [INITIAL_SEROPREVALENCE [INITIAL_SEROPREVALENCE ...]]
Seroprevalence of the population (default to zero). The seroprevalence should always be
greater than or euqal to initial incidence rate. The difference between seroprevalence and
incidence rate will determine the "recovered" rate of the population. Multipliers can be
specified to set incidence rate of for particular groups (e.g. --initial-incidence-rate 0.1
docter=1.2 will set incidence rate to 0.12 for doctors).
--infectors [INFECTORS [INFECTORS ...]]
Infectees to introduce to the population, default to '0'. If you would like to introduce
multiple infectees to the population, or if you have named groups, you will have to specify
the IDs of carrier such as --infectors nurse1 nurse2. Specifying this parameter without
value will not introduce any infector.
--interval INTERVAL Interval of simulation, default to 1/24, by hour
--logfile LOGFILE logfile
--prop-asym-carriers [PROP_ASYM_CARRIERS [PROP_ASYM_CARRIERS ...]]
Proportion of asymptomatic cases. You can specify a fix number, or two numbers as the lower
and higher CI (95%) of the proportion. Default to 0.10 to 0.40. Multipliers can be
specified to set proportion of asymptomatic carriers for particular groups.
--stop-if [STOP_IF [STOP_IF ...]]
Condition at which the simulation will end. By default the simulation stops when all
individuals are affected or all infected individuals are removed. Current you can specify a
time after which the simulation will stop in the format of `--stop-if "t>10"' (for 10
days).
--allow-lead-time The seed carrier will be asumptomatic but always be at the beginning of incurbation time.
If allow lead time is set to True, the carrier will be anywhere in his or her incubation
period.
--plugin ... One or more of "--plugin MODULE.PLUGIN [args]" to specify one or more plugins. FLUGIN will
be assumed to be MODULE name if left unspecified. Each plugin has its own parser and can
parse its own args.
-j JOBS, --jobs JOBS Number of process to use for simulation. Default to number of CPU cores.
-s STAT_INTERVAL, --stat-interval STAT_INTERVAL
Interval for statistics to be calculated, default to 1. No STAT event will happen it it is
set to 0.
Example usages¶
Homogeneous and heterogeneous populations¶
outbreak_simulator
simulates the outbreak of COVID-19 in a population with 64 individuals, with one introduced infector.
outbreak_simulator --popsize nurse=10 patient=100 --infector patient0
simulates a population with 10 nurses and 100 patients when the first patient
carries the virus.
Change number of infectors¶
outbreak_simulator --infector 0 1 --pre-quarantine 7 0 1
simulates the introduction of two infectors, both after 7 days of quarantine. Here
0 and 1 are IDs of individuals
Changing model parameters¶
outbreak_simulator --prop-asym-carriers 0.10
runs the simulation with a fixed ratio of asymptomatic carriers.
outbreak_simulator --incubation-period normal 4 2
runs the simulation incubation period sampled from a normal distribution with mean 4 and standard deviation of 2.
Specigy group-specific parameters¶
Parameters symptomatic-r0, asymptomatic-r0 and incubation-period can be
set to different values for each groups. These are achived by “multipliers”,
which multiplies specified values to values drawn from the default distribution.
For example, if in a hospital environment nurses, once affected, tends to have
higher R0 because he or she contact more patients, and on the other hand
patients are less mobile and should have lower R0. In some cases the nurses
are even less protected and are more susceptible. You can run a simulation
with two patients carrying the virus with the following options:
outbreak_simulator --popsize nurse=10 patient=100 \
--symptomatic-r0 nurse=1.5 patient=0.8 \
--asymptomatic-r0 nurse=1.5 patient=0.8 \
--susceptibility nurse=1.2 patient=0.8 \
--infector patient0 patient1
Plug-in system (advanced usages)¶
It is very likely that for complex simulations you would have scenarios that are not provided by the core simulator. This simulator has a plug-in system that allows you to define your own functions that would be called by the simulator at specified intervals or events.
Specify one or more plugins from command line¶
To use plugins for a simulation, they should be specified as:
outbreak_simulator \
# regular parameters
-j 4 \
# first system plugin
--plugin contact_tracing --succ-rate 0.8 \
# second (customied) plugin
--plugin modulename1.plugin_name1 --start 10 --par2 val2 --par3
where
- Each plugin is defined after a
--pluginparameter and before another--pluginor the end of command line arguments. - A plugin can be a system plugin that is located under the
pluginsdirectory, or a cutomized plugin that you have defined. - A plugin can be specified by
modulenameif it is the name of the python module and the name of the plugin class, ormodulename.plugin_nameif the class has a different name. - Plugins accept a common set of parameters (e.g.
--start,--end) to define when the plugin will be triggered. - The rest of the plugin parameters will be parsed by the plugin
System plugins¶
Common paramters of plugins¶
% outbreak_simulator --plugin -h
A plugin for covid19-outbreak-simulator
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
Plugin init¶
% outbreak_simulator --plugin init -h
usage: --plugin init [-h] [--start START] [--end END] [--at AT [AT ...]] [--interval INTERVAL]
[--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]] [--incidence-rate [INCIDENCE_RATE [INCIDENCE_RATE ...]]]
[--seroprevalence [SEROPREVALENCE [SEROPREVALENCE ...]]]
Initialize population with initial prevalence and seroprevalence
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
--incidence-rate [INCIDENCE_RATE [INCIDENCE_RATE ...]]
Incidence rate of the population (default to zero), which should be the probability that any individual is
currently affected with the virus (not necessarily show any symptom). Multipliers can be specified to set
incidence rate of for particular groups (e.g. --initial-incidence-rate 0.1 docter=1.2 will set incidence rate
to 0.12 for doctors).
--seroprevalence [SEROPREVALENCE [SEROPREVALENCE ...]]
Seroprevalence of the population (default to zero). The seroprevalence should always be greater than or euqal
to initial incidence rate. The difference between seroprevalence and incidence rate will determine the
"recovered" rate of the population. Multipliers can be specified to set incidence rate of for particular
groups (e.g. --initial-incidence-rate 0.1 docter=1.2 will set incidence rate to 0.12 for doctors).
Plugin dynamic-r0¶
% outbreak_simulator --plugin dynamic-r0 -h
usage: --plugin dynamic_r0 [-h] [--start START] [--end END] [--at AT [AT ...]] [--interval INTERVAL]
[--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]]
[--new-symptomatic-r0 NEW_SYMPTOMATIC_R0 [NEW_SYMPTOMATIC_R0 ...]]
[--new-asymptomatic-r0 NEW_ASYMPTOMATIC_R0 [NEW_ASYMPTOMATIC_R0 ...]]
Change multiplier number at specific time.
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
--new-symptomatic-r0 NEW_SYMPTOMATIC_R0 [NEW_SYMPTOMATIC_R0 ...]
Updated production number of symptomatic infectors, should be specified as a single fixed number, or a range,
and/or multipliers for different groups such as A=1.2. For example "--symptomatic-r0 1.4 2.8 nurse=1.2" means
a general R0 ranging from 1.4 to 2.8, while nursed has a range from 1.4*1.2 and 2.8*1.2.
--new-asymptomatic-r0 NEW_ASYMPTOMATIC_R0 [NEW_ASYMPTOMATIC_R0 ...]
Updated production number of asymptomatic infectors, should be specified as a single fixed number, or a range
and/or multipliers for different groups
Plugin stat¶
% outbreak_simulator --plugin stat -h
usage: --plugin stat [-h] [--start START] [--end END] [--at AT [AT ...]] [--interval INTERVAL]
[--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]]
Print STAT information
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
Plugin sample¶
% outbreak_simulator --plugin sample -h
usage: --plugin sample [-h] [--start START] [--end END] [--at AT [AT ...]] [--interval INTERVAL]
[--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]] (--proportion PROPORTION | --size SIZE)
Draw random sample from the population
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
--proportion PROPORTION
Proprotion of the population to sample.
--size SIZE Number of individuals to sample.
Plugin quarantine¶
% outbreak_simulator --plugin quarantine -h
usage: --plugin quarantine [-h] [--start START] [--end END] [--at AT [AT ...]] [--interval INTERVAL]
[--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]] [--proportion PROPORTION] [--duration DURATION]
[IDs [IDs ...]]
Quarantine specified or all infected individuals for specified durations.
positional arguments:
IDs IDs of individuals to quarantine.
optional arguments:
-h, --help show this help message and exit
--start START Start time. Default to 0 no parameter is defined so the plugin will be called once at the beginning.
--end END End time, default to none, meaning there is no end time.
--at AT [AT ...] Specific time at which the plugin is applied.
--interval INTERVAL Interval at which plugin is applied, it will assume a 0 starting time if --start is left unspecified.
--trigger-by [TRIGGER_BY [TRIGGER_BY ...]]
Events that triggers this plug in.
--proportion PROPORTION
Proportion of affected individuals to quarantine. Default to all, but can be set to a lower value to indicate
incomplete quarantine. This option does not apply to cases when IDs are explicitly specified.
--duration DURATION Days of quarantine
Implementation of plugins¶
Implementation wise, the plugins should be written as Python classes that
- Drive from
outbreak_simulator.BasePluginand calls its__init__function. This will pointself.simulator, andself.loggerof the plugin to the simulator, population, and logger of the simulator.simulatorhas properties such assimu_args,params, andmodelso you can access command line simulator arguments, model parameters, and change simulation model (e.g. parameters). - Provide as member function:
get_parser()(optional): a function that extends the base parser to add plugin specific arguments.apply(time, population, args)(required): A function that accepts parameterstime(time when the plugin is called) andargs(plugin args).
The plugin can change the status of the population or simulation parameters, write to system log simulator.logger or write to its own output files. It should return a list of events that can happen at a later time, or an empty list indicating no future event is triggered.
The base class will handle the logics when the plugin will be executed (parameters --start, --end, --interval
--at and --trigger-by). Please check the plugins directory for examples on how to write plugins for the simulator.
Output from the simulator¶
Events tracked during the simulation¶
The output file contains events that happens during the simulations. For example, for command
outbreak_simulator --repeat 100 --popsize 64 --logfile result_remove_symptomatic.txt
You will get an output file result_remove_symptomatic.txt with the following columns:
| column | content |
|---|---|
id |
id of the simulation. |
time |
time of the event in days, accurate to hour. |
event |
type of event |
target |
subject of the event, for example the ID of the individual that has been quarantined. |
params |
Additional parameters, mostly for the INFECTION event where simulated $R_0$ and incubation period will be displayed. |
Currently the following events are tracked
| Name | Event |
|---|---|
INFECTION |
Infect an non-quarantined individual, who might already been infected. |
INFECION_FAILED |
No one left to infect |
INFECTION_AVOIDED |
An infection happended during quarantine. The individual might not have showed sympton. |
INFECTION_IGNORED |
Infect an infected individual, which does not change anything. |
SHOW_SYMPTOM |
Show symptom. |
REMOVAL |
Remove from population. |
RECOVER |
Recovered, no longer infectious |
QUARANTINE |
Quarantine someone till specified time. |
REINTEGRATION |
Reintroduce the quarantined individual to group. |
STAT |
Report population stats at specified intervals. |
ABORT |
If the first carrier show sympton during quarantine. |
END |
Simulation ends. |
ERROR |
An error was raised. |
The log file of a typical simulation would look like the following:
# CMD: outbreak_simulator --rep 3 --plugin stat --interval 1
# START: 06/21/2020, 00:02:06
id time event target params
2 0.00 INFECTION 0 leadtime=2.26,r0=1.94,r=0,r_presym=0,r_sym=0,incu=3.07
2 0.00 STAT . n_recovered=0,n_infected=1,n_popsize=64,incidence_rate=0.0156,seroprevalence=0.0156
2 0.81 SHOW_SYMPTOM 0 .
2 0.81 REMOVAL 0 popsize=63
2 7.79 RECOVER 0 recovered=0,infected=0,popsize=63,True=True
2 7.79 END 63 popsize=63,prop_asym=0.288
3 0.00 INFECTION 0 leadtime=4.82,r0=2.20,r=0,r_presym=0,r_sym=0,incu=4.82
3 0.00 STAT . n_recovered=0,n_infected=1,n_popsize=64,incidence_rate=0.0156,seroprevalence=0.0156
3 0.00 SHOW_SYMPTOM 0 .
3 0.00 REMOVAL 0 popsize=63
3 6.40 RECOVER 0 recovered=0,infected=0,popsize=63,True=True
3 6.40 END 63 popsize=63,prop_asym=0.260
1 0.00 INFECTION 0 leadtime=0.16,r0=2.49,r=1,r_presym=1,r_sym=0,incu=2.38
1 0.00 STAT . n_recovered=0,n_infected=1,n_popsize=64,incidence_rate=0.0156,seroprevalence=0.0156
1 2.14 INFECTION 20 by=0,r0=0.45,r=1,r_asym=1
1 2.22 SHOW_SYMPTOM 0 .
1 2.22 REMOVAL 0 popsize=63
1 6.61 INFECTION 36 by=20,r0=1.69,r=1,r_presym=1,r_sym=0,incu=7.04
1 9.43 RECOVER 0 recovered=0,infected=2,popsize=63,True=True
1 10.55 INFECTION 19 by=36,r0=0.49,r=1,r_asym=1
1 13.39 INFECTION 62 by=19,r0=1.44,r=0,r_presym=0,r_sym=0,incu=3.65
1 13.65 SHOW_SYMPTOM 36 .
1 13.65 REMOVAL 36 popsize=62
1 14.14 RECOVER 20 recovered=1,infected=3,popsize=62
1 17.04 SHOW_SYMPTOM 62 .
1 17.04 REMOVAL 62 popsize=61
1 19.30 RECOVER 36 recovered=1,infected=2,popsize=61,True=True
1 22.55 RECOVER 19 recovered=2,infected=2,popsize=61
1 23.83 RECOVER 62 recovered=2,infected=2,popsize=61,True=True
1 23.83 END 61 popsize=61,prop_asym=0.092
# START: 06/21/2020, 00:02:07
which I assume would be pretty self-explanatory. Note that the simulation IDs are not ordered because the they are run in parallel but you can expect all events belong to the same simulation are recorded together..
Summary report from multiple replicates¶
At the end of each command, a report will be given to summarize key statistics from multiple replicated simulations. The output contains the following keys and their values
| name | value |
|---|---|
logfile |
Log file of the simulation with all the events |
popsize |
Initial population size |
handle_symptomatic |
If asymptomatic infectees are kept, removed, or quarantined, at what percentage. |
prop_asym_carriers |
Proportion of asymptomatic carriers, also the probability of infectee who do not show any symptom |
interval |
Interval of time events (1/24 for hours) |
n_simulation |
Total number of simulations, which is the number of END events |
total_infection |
Number of INFECTION events |
total_infection_failed |
Number of INFECTION_FAILED events |
total_infection_avoided |
Number of INFECTION_AVOIDED events |
total_infection_ignored |
Number of INFECTION_IGNORED events |
total_show_symptom |
Number of SHOW_SYMPTOM events |
total_removal |
Number of REMOVAL events |
total_quarantine |
Number of QUARANTINE events |
total_reintegration |
Number of REINTEGRATION events |
total_abort |
Number of ABORT events |
total_asym_infection |
Number of asymptomatic infections |
total_presym_infection |
Number of presymptomatic infections |
total_sym_infection |
Number of symptomatic infections |
n_remaining_popsize_XXX |
Number of simulations with XXX remaining population size |
n_no_outbreak |
Number of simulations with no outbreak (no symptom from anyone, or mission canceled) |
n_outbreak_duration_XXX |
Number of simulations with outbreak ends in day XXX. Pre-quarantine days are not counted as outbreak. Outbreak can end at day 0 if the infectee will not show symtom or infect others. |
n_no_infected_by_seed |
Number of simulations when the introduced carrier does not infect anyone |
n_num_infected_by_seed_XXX |
Number of simulations with XXX people affected by the introduced virus carrier, XXX > 0 . |
n_first_infected_by_seed_on_day_XXX |
Number of simulations when the introduced carrier infect the first infectee on day XXX, XXX<1 is rounded to 1, and so on. Pre-quarantine time is deducted. |
n_seed_show_no_symptom |
Number of simulations when the seed show no symptom |
n_seed_show_symptom_on_day_XXX |
Number of simulations when the carrier show symptom at day XXX, XXX < 1 is rounded to 1, and so on. |
n_no_first_infection |
Number of simualations with no infection at all. |
n_first_infection_on_day_XXX |
Number of simualations with the first infection event happens at day XXX. It is the same as XXX_n_first_infected_by_seed_on_day but is reserved when multiple seeds are introduced. |
n_first_symptom |
Number of simulations when with at least one symptomatic case |
n_first_symptom_on_day_XXX |
Number of simulations when the first symptom appear at day XXX, XXX < 1 is rounded to 1, and so on. Symptom during quarantine is not considered and pre-quarantine days are deducted. |
n_second_symptom |
Number of simulations when there are a second symptomatic case symptom. |
n_second_symptom_on_day_XXX |
Number of simulations when the second symptom appear at day XXX after the first symptom |
n_third_symptom |
Number of simulations when there are a third symptomatic case symtom |
n_third_symptom_on_day_XXX |
Number of simulations when the first symptom appear at day XXX after the second symptom |
n_popsize_XXX |
Population size at time XXX, a list if multiple replicates |
n_YYY_popsize_XXX |
Population size of group YYY at time XXX a list if multiple replicates |
n_infected_XXX |
Number of infected individuals at time XXX a list if multiple replicates |
n_YYY_infected_XXX |
Number of infected individuals in group YYY at time XXX a list if multiple replicates |
n_recovered_XXX |
Number of recovered individuals at time XXX a list if multiple replicates |
n_YYY_recovered_XXX |
Number of recovered individuals in group YYY at time XXX a list if multiple replicates |
incidence_rate_XXX |
Incidence rate (infected/popsize) at time XXX a list if multiple replicates |
YYY_incidence_rate_XXX |
Incidence rate in group YYY at time XXX a list if multiple replicates |
seroprevalence_XXX |
Seroprevalence (infected + recovered)/popsize) at time XXX a list if multiple replicates |
YYY_seroprevalence_XXX |
Seroprevalence in group YYY at time XXX a list if multiple replicates |
avg_n_popsize_XXX |
Average population size at time XXX if there are multiple replicates |
avg_n_YYY_popsize_XXX |
Average population size of group YYY at time XXX if there are multiple replicates |
avg_n_infected_XXX |
Average number of infected individuals at time XXX if there are multiple replicates |
avg_n_YYY_infected_XXX |
Average number of infected individuals in group YYY at time XXX if there are multiple replicates |
avg_n_recovered_XXX |
Average number of recovered individuals at time XXX if there are multiple replicates |
avg_n_YYY_recovered_XXX |
Average number of recovered individuals in group YYY at time XXX if there are multiple replicates |
avg_incidence_rate_XXX |
Average incidence rate (infected/popsize) at time XXX if there are multiple replicates |
avg_YYY_incidence_rate_XXX |
Average incidence rate in group YYY at time XXX if there are multiple replicates |
avg_seroprevalence_XXX |
Average neroprevalence (infected + recovered)/popsize) at time XXX if there are multiple replicates |
avg_YYY_seroprevalence_XXX |
Average seroprevalence in group YYY at time XXX if there are multiple replicates |
EVENT_STAT_XXX |
Reported statistics STAT for customized event EVENT at time XXX a list if multiple replicates |
avg_EVENT_STAT_XXX |
Average reported statistics STAT for customized event EVENT at time XXX if there are multiple replicates |
Data analysis tools¶
Because all the events have been recorded in the log files, it should not be too difficult for
you to write your own script (e.g. in R) to analyze them and produce nice figures. We however
made a small number of tools available. Please feel free to submit or own script for inclusion in the contrib
library.
time_vs_size.R¶
The ``contrib/time_vs_size.R` <https://github.com/ictr/covid19-outbreak-simulator/blob/master/contrib/time_vs_size.R>`_ script provides an example on how to process the data and produce a figure. It can be used as follows:
Rscript time_vs_size.R simulation.log 'COVID19 Outbreak Simulation with Default Paramters' time_vs_size.png
and produces a figure
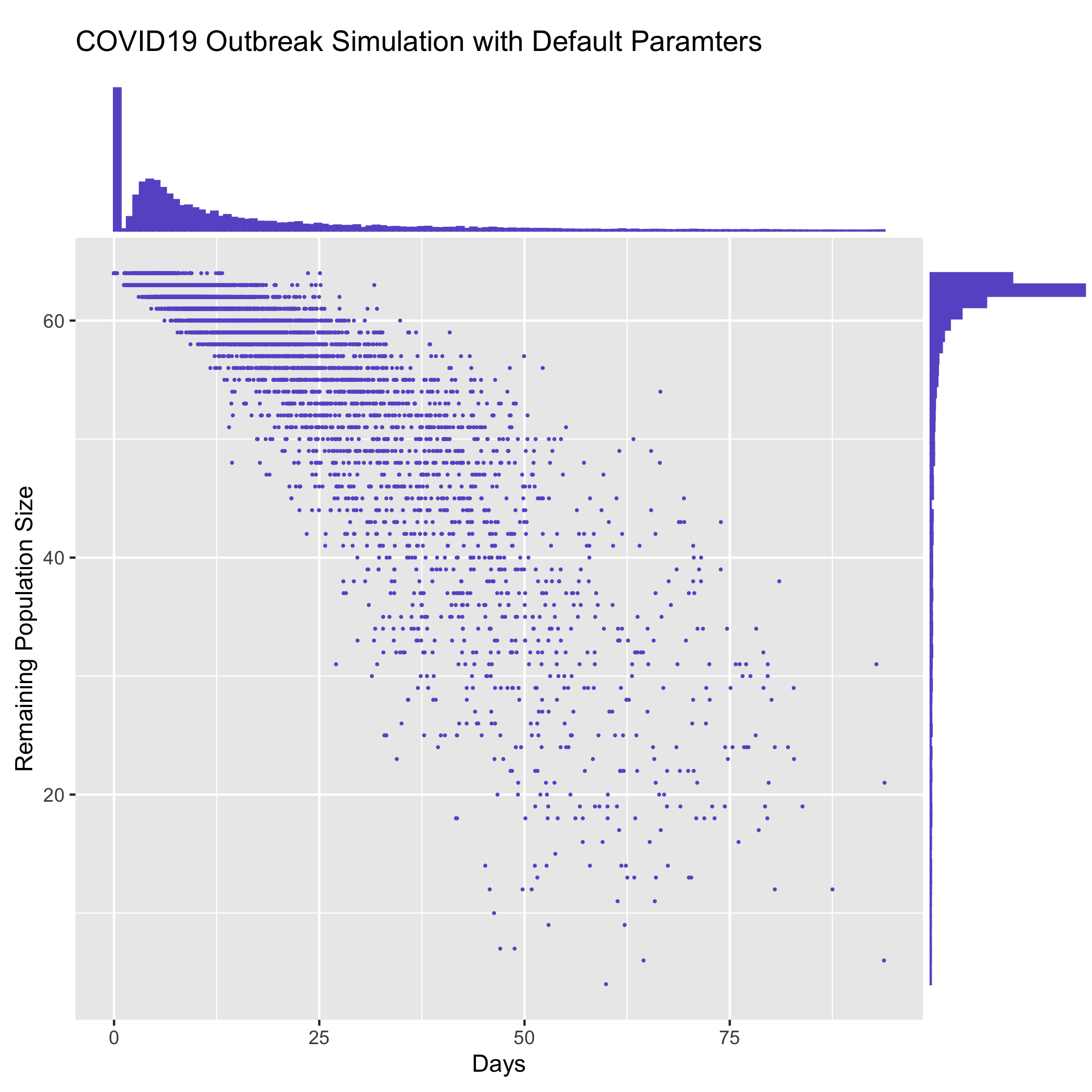
merge_summary.py¶
``contrib/merge_summary.py` <https://github.com/ictr/covid19-outbreak-simulator/blob/master/contrib/merge_summary.py>`_ is a script to merge summary stats from multiple simulation runs.
Acknowledgements¶
This tool has been developed and maintained by Dr. Bo Peng, associate professor at the Baylor College of Medicine, with guidance from Dr. Christopher Amos, from the Institute for Clinical and Translational Research, Baylor College of Medicine. Contributions to this project are welcome. Please refer to the LICENSE file for proper use and distribution of this tool.
This package was created with Cookiecutter and the ``audreyr/cookiecutter-pypackage` <https://github.com/audreyr/cookiecutter-pypackage>`_ project template.